Inputs
*Note: Input files all should be put under user-defined directories, which we call execution directories, for execution of cisMuton, cisFusion, and cisCton. Execution directories for cisMuton, cisFusion, and cisCton must be separately prepared.
1. User-Prepared Data
Here we assume the following files as user-prepared data:
- .fastq files
- .bed files:
- Target-capture regions
- Regions containing the breakpoints of fusion genes
For other input files, see the cisCall Installation section.
1.1. .fastq Files
The .fastq files to be recognized by cisCall should be named:
- For pared-end read files:
- sample .fastq files:
${SAMPLE_NAME}.${GROUP}.fastq.R1.gz,${SAMPLE_NAME}.${GROUP}.fastq.R2.gz. - control (background) .fastq files:
${BG_SAMPLE_NAME}.${GROUP}.fastq.R1.gz,${BG_SAMPLE_NAME}.${GROUP}.fastq.R2.gz.
- sample .fastq files:
- For single-end read files:
- sample .fastq file:
${SAMPLE_NAME}.${GROUP}.fastq.gz - control (background) .fastq file:
${BG_SAMPLE_NAME}.${GROUP}.fastq.gz
- sample .fastq file:
Here, ${GROUP} is a group name to identify a group of input files in the execution directory.
Please assign the same group name to ${GROUP} across all input files.
The same group name is used between cisMuton and cisCton.
A different group name can be used for cisFusion.
1.2. .bed files
All cisCall modules require .bed files as input data:
${GROUP}.target.bed/${GROUP}.bed: target-capture regions.${GROUP}.fusion.bed: regions containing the breakpoints of fusion genes.
See the cisCall Installation section for details.
1.2.1. Target-Capture Regions
- cisMuton/cisFusion:
${GROUP}.target.bed - cisCton:
${GROUP}.bed
These have nearly the same format. See the cisCall installation section for details.
1.2.2. Regions Containing the Breakpoints of Fusion Genes
- cisMuton/cisFusion:
${GROUP}.fusion.bed - cisCton:
target_fusion.txt. The contents are the same as in${GROUP}.fusion.bed, though the name is slightly different.
2. Setting Parameters
Parameters must be set for cisMuton/cisFusion depending on experimental conditions (e.g., frozen/cell-line or FFPE samples) and sequencers (Illumina or Ion sequencer).
Please set these for cisMuton and cisFusion separately, since the cisMuton and cisFusion commands are different despite using the same name (qc_run.pl).
*Note: The current version of cisCton only assumes FFPE samples and an Illumina sequencer, so the parameters do not need to be set.
Change the current directory to the cisMuton/cisFusion execution directory and issue the command.
If theparam/directory already exists in the execution directory, delete or rename it beforehand.For cisMuton,
$ perl /home/cisusr/cisCall/cisMuton/bin/qc_run.pl ::${GROUP}:PARAMFor cisFusion,
$ perl /home/cisusr/cisCall/cisFusion/bin/qc_run.pl ::${GROUP}:PARAMA pop-up window will appear for setting the parameters.
In the Select condition window, select an item based on the sequencing platform and sample type.
- There are 11 choices:
- Options 7 and 8: tuned well
- Options 1 and 4: tuned to some degree
- Options 9 and 10: less tuned
- Other options: under trial
"Select condition" window

- There are 11 choices:
When the Edit Parameter window (see the image bellow) appears, select 2: Save.
"Edit parameter" window
When the Save param-ID window (see image below) appears, select 0: To be automatically named.
"Save param-ID" window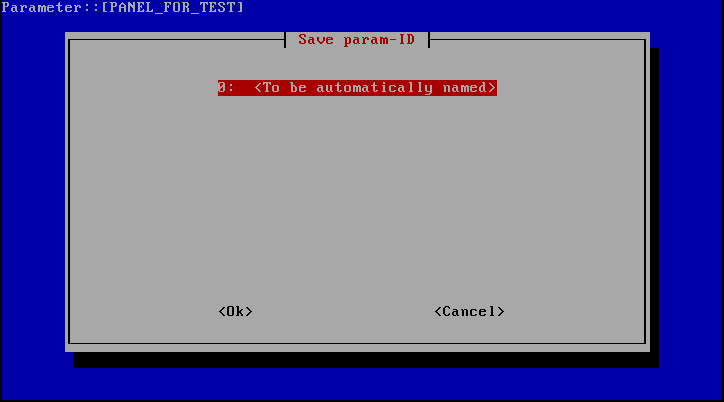
On the Edit Parameter window, select 1: Quit. The pop-up window then disappears.
Check if the parameter file
param/${GROUP}.0.txtwas generated under the execution directory.
*Note: This file should be edited to change the paths to BWA (bwa) or Samtools (samtools) if you use self-compiled commands.